Arnaba Saha Chaity, Dipa Roy, Tamanna Nasrin and Ariful Haque from the different institute of the Rajshahi. wrote a research article about, Antibiotic Resistance in Multidrug-Resistant Bacteria: Bangladesh Study. entitled, Antibiotic Resistance and Integron Prevalence among Multidrug-Resistant Bacterial in Bangladesh. Molecular Pathology Laboratory, Institute of Biological Sciences, University of Rajshahi, Rajshahi-6205, Bangladesh. This research paper published by the International Journal of Biosciences | IJB. an open access scholarly research journal on Biology, under the affiliation of the International Network For Natural Sciences | INNSpub. an open access multidisciplinary research journal publisher.
Abstract
Antibiotic-resistant
bacterial strains are widespread in hospitals and intensive care units. This
poses a serious threat to human health as the effectiveness of many antibiotics
has been diminished by the emergence of resistant strains. The overuse of
β-lactam antibiotics has led to the rise of antibiotic-resistant bacteria,
including Extended-Spectrum β-Lactamase (ESBL) producing strains. However, ESBL
screening is not commonly performed in Bangladesh, despite the growing
prevalence of antibiotic resistance. Multidrug-resistant strains, particularly
those carrying the Integron integrase 1 gene is responsible for antibiotic
resistance. Horizontal integron transfer is one of the key factors that can
contribute to the emergence of multidrug-resistant (MDR) bacteria. In
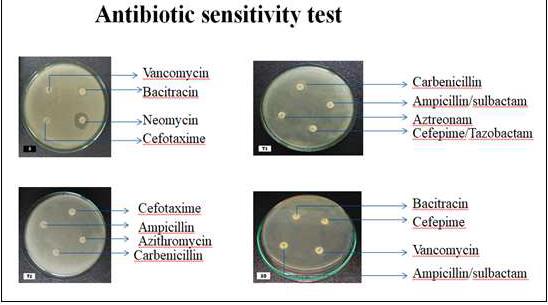
this study, antibiotic sensitivity tests were conducted using 25 antibiotics.
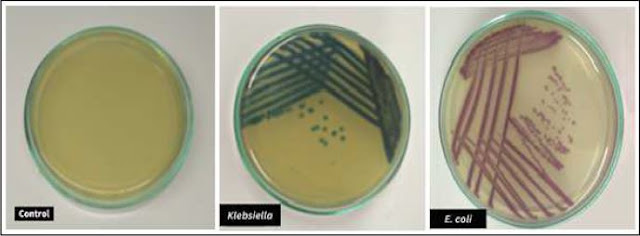
It was found that E. coli and Klebsiella both showed
resistance to Aztreonam, Ampicillin/Sulbactam, Amoxyclav, Cefepime,
Cefepime/Tazobactam, Ampicillin and Cefotaxime antibiotics. Our findings
suggest that integron is common among MDR isolates and that they can be
used to identify MDR isolates. As a result of the possibility of a widespread
outbreak of MDR isolates, molecular surveillance and integron sequencing in
other parts of the country is advised. The purpose of this study is innovation
to create new antibiotics and alternative treatments to address
antibiotic-resistant bacteria.
Read more : Oyster Nut:Ethnobotanical Insights from Northern Tanzania | Informativebd
Introduction
Antimicrobial resistance poses a growing threat to human health and medical treatments and rising multidrug-resistant nosocomial infections in hospitals. Prompt treatment is crucial for vulnerable individuals at risk of bacterial infections. Viral infections are more likely to be the source of localized redness, swelling, and discomfort (Murray et al., 1998).
Antimicrobial resistance in bacteria is a complicated process involving a variety of different mechanisms. Susceptible bacteria can develop resistance through mutations or the transfer of resistance genes found on mobile DNA elements like integron (Barlow et al. 2004, Normak & Normak 2002). This, integron play a crucial role in the spread of antibiotic resistance, especially in Gram-negative bacteria by attaching to mobile genetic elements like transposons and conjugative plasmids, resistance integron transmit antibiotic resistance. (Mazel, 2006). Integron share antibiotic resistance gene cassettes predominantly through convergent evolution. Mobile integron has had a considerable impact on the spread of antibiotic resistance, particularly in Gram-negative bacteria (Davies et al., 2010). From the pool of these environmental genetic elements that is available, Integron has amassed a wide variety of resistance genes. Furthermore, their prevalence has significantly increased, increasing the possibility of interactions with other DNA and the emergence of additional, more complex mobile elements that are resistant to different antibiotic classes, disinfectants, and heavy metals (Gillings & Stokes 2012).
Class 1 integron are major contributors to antibiotic resistance in Gram-negative bacteria like Klebsiella and E. coli. They evolve due to exposure to selection agents in human-populated areas and natural environments, acquiring advantageous genes. The presence of resistance gene cassettes within class 1 integron is linked to antibiotic resistance. Addressing this problem requires essential research and intervention to combat the spread of antibioticresistant microorganisms. Ongoing development and dissemination of antibiotic resistance in Gramnegative bacteria like Klebsiella and E. coli class 1 integron play a crucial role. Their evolution is fueled by persistent exposure to selection agents in both human-populated areas and natural environments, leading to the acquisition of beneficial genes. The presence of resistance gene cassettes within class 1 integron is closely associated with antibiotic resistance. To effectively combat antibiotic-resistant microorganisms, it is essential to prioritize continuous research and innovation. Class 1 integrons are commonly found in various Gram-negative bacteria such as Acinetobacter, Vibrio, Aeromonas, Proteus, Burkholderia, Alcaligenes, Campylobacter, Enterobacter, Citrobacter, Klebsiella, Mycobacterium, Pseudomonas, Serratia, Salmonella, Shigella, and Escherichia coli (Yu G, Li Y and Liu X. 2013). Several studies have investigated prevalence of integron in MDR Escherichia coli and Klebsiella isolates around the world. These studies have identified a significant link between the presence of integrons and antibiotic resistance.
The study of integron aims to effectively combat antibiotic resistance by developing precise drugs that can be tailored to individual patients at a low cost, resulting in faster recovery times. With the emergence of antibiotic-resistant bacteria posing a significant threat to public health, there is a growing global interest in exploring integron profiles for the design of novel drugs that can safeguard both human and economically valuable animal populations. By understanding the intricate mechanisms of integron, scientists are hopeful of discovering innovative drugs that can tackle antibiotic resistance, thereby protecting human health and ensuring the well-being of economically important animals. This renewed focus on integron holds the potential to revolutionize the field of medicine and pave the way for personalized and cost-effective antibiotic treatments.
Reference
Barlow RS, Pemberton
JM, Desmarchelier PM, Gobius KS. 2004. Isolation and characterization of
integron-containing bacteria without antibiotic selection. Antimicrobial
agents and chemotherapy 48(3), 838-842.
Davies, Julian, Dorothy
Davies. 2010.”Origins and evolution of antibiotic resistance.”
Microbiology and molecular biology reviews 74(3), 417-433.
Gillings MR, Stokes HW. 2012.
Are humans increasing bacterial evolvability?. Trends in ecology &
evolution 27(6), 346-352.
Mazel D. 2006.
Integrons: agents of bacterial evolution. Nature Reviews
Microbiology 4(8), 608-620.
Murray AE, Chambers JJ,
Van Saene HK. 1998. Infections in patients requiring ventilation in
intensive care: application of a new classification. Clinical microbiology
and infection 4(2), 94-99.
Normark BH, Normark S. 2002.
Evolution and spread of antibiotic resistance. Journal of internal
medicine 252(2), 91-106.
Saha AK, Haque MF,
Karmaker S, Mohanta MK. 2009. Antibacterial effects of some antiseptics
and disinfectants. Journal of Life and Earth Science, 19-21.
Yu G, Li Y, Liu X. 2013.
Role of integrons in antimicrobial resistance: A review. African Journal
of Microbiology Research Apr; 7(15), 1301–1310.










%20in%20full.JPG)


0 comments:
Post a Comment