K. Archana Rao, S. Sangeetha and SA. Lakshminarayana from the different institute of the india, wrote a research article about Detecting NDM-1 Gene in MBL-Producing Pseudomonas aeruginosa, entitled, "NDM-1 gene detection from Metallobeta lactamase (MBL) producing Pseudomonas aeruginosa: A pilot study from a teritiary care centre". This research paper published by the International Journal of Microbiology and Micology | IJMM, an open access scholarly research journal on Microbiology, under the affiliation of the International Network For Natural Sciences |INNSpub, an open access multidisciplinary research journal publisher.
Abstract
Multidrug resistant
bacteria always remain a great challenge. The latest threat being New Delhi
Metallobetalactamase-1 (NDM-1) a superbug has brought notoriety to Indian
Health care. NDM-1 refers to the transmissible genetic element encoding
multiple resistant genes, first isolated from a strain of Klebsiella spp.
in New Delhi, India, which has the ability to hydrolyse beta lactams and
carbapenams. Detection of NDM-1 gene in multidrug resistant Pseudomonas
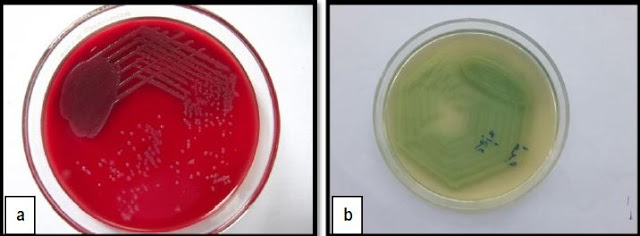
isolates from various clinical samples. 200 Pseudomonas species were isolated
in Microbiology laboratory during one year period were included in the study.
Samples were processed as per Standard operating procedures. Antibiotic
sensitivity testing was done by Kirby-Bauer disc diffusion method. The results were
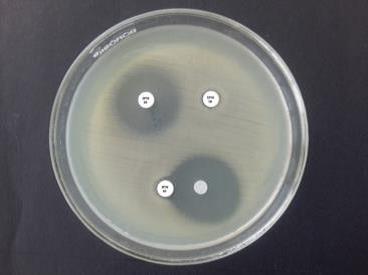
interpreted as per CLSI guidelines. MBL detection was done, by using EDTA
Double Disc Synergy Test and Imipenem [I]-EDTA Combined Disc Test. MBL positive
isolates were subjected to conventional PCR for genotyping & detection of
NDM-1. A cross sectional descriptive study. Out of 12545 samples that were
received in microbiology laboratory, 299 Non-Fermenting Gram Negative Bacilli
[NFGNB] were isolated of which 200 were speciated as Pseudomonas aeruginosa. 20/200 [10%] were resistant to imipenem and 24/200 [12%] to
meropenem. 10% of isolates showed MBL positive. NDM-1 gene was not detected in
any of the 20 MBL positive isolates. NDM-1 gene since its origin has caused
chaos in the health care facility with its ability to cause various infections.
Detection is possible only with molecular methods. Thus gene detection plays a
pivotal role in patient treatment and reduction of hospital stay. (Pseudomonas,
multidrug resistant, New Delhi Metallobetalactamase-1, super bugs).
Read more : Green Synthesis: Silver Nanoparticles from Moringa oleifera | InformativeBD
Introduction
Pseudomonas aeruginosa a gram-negative bacterium is one of the leading causes of health care associated infections. Multi-drug-resistant Pseudomonas aeruginosa is a growing concern. Multi drug resistant bacteria are defined as isolates that show intermediate or resistance to at least three drugs in the following classes: beta-lactams, carbapenems, aminoglycosides, and fluoroquinolones.(1) Reported rates of multi drug resistant Pseudomonas aeruginosa varies from 0.6-32% based on geographic location and type of surveillance study.(2) Pseudomonas aeruginosa is intrinsically resistant to a wide range of antibiotics like ampicillin, cefuroxime and cefotaxim which is attributed to its production of ß-lactamases.(3) Indiscriminate use of antibiotics, heavy antibiotic pressure further accentuates the mutations in genes coding for ß-lactamase enzymes. This results in the fabrication of new ß-lactamases with wider ranges of activity. The emergence of New Delhi Metallobetalactamase-1 (NDM-1) Pseudomonas aeruginosa, a superbug, is a potential threat to human health. Among clinically significant carbapenamases, NDM-1 is the biggest menace. It significantly hydrolyses beta-lactams and carbapenems. NDM-1 producing strains exhibit multidrug resistant profile because they also harbor genes that encode for resistance to aminoglycosides and fluoroquinolones.(4)(5)
This mixed bag of rapidly emerging antimicrobial resistant organisms and increasing rates of healthcare infections has always drawn the attention of clinical microbiologists and thus put us under an obligation to detect these resistance mechanism at the earliest. Emerging ‘Superbugs or Multi-Drug Resistant (MDR)’ pathogens have always been an enduring hitch in the health care settings and also challenges the effectiveness and usefulness of even most potent antibiotics. (6)Knowledge of NDM and its prevalence is essential because P. aeruginosa with intrinsic colonization capacity has the ability to persist in the hospital environment for indefinite periods but there is paucity of such reports. Hence the present study was undertaken as a pilot project to detect the NDM-1 gene in multi drug resistant Pseudomonas species, because determination of resistance mechanism helps to formulate efficient antibiotic policy and infection control protocols for holistic health care.
Objectives
1) Identification of
pseudomonas species from various clinical isolates
2) Detection of their
antimicrobial resistance
3) Detection of MBL
production by screening tests
4) Detection of NDM-1 gene in the MBL positive isolates
Reference
Ahuja AS, Kabra SK. 2002.
“Cystic fibrosis: Indian experience.” Ind Pediatr 39, 813-818.
Clinical and Laboratory
Standards Institute. 2018. Performance Standards for Antimicrobial Disks
Susceptibility Tests; Approved Standard. 10th ed. CLSI document M02-A10. Wayne,
PA: CLSI.
Deshpande P, Rodrigues
C, Shetty A. 2010. New Delhi Metallo beta lactamase (NDM-1) in
Enterobacteriaceae : treatment options with carbapenems compromised. J Assoc
Physicians India 58, 147–9.
Falagas ME, Koletsi PK,
Bliziotis IA. 2006. The diversity of definitions of multidrug-resistant
(MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas
aeruginosa. J Med Microbiol 55(Pt 12), 1619-29.
Fazeli H1, Akbari R,
Moghim S, Narimani T, Arabestani MR, Ghoddousi AR. 2012. Pseudomonas
aeruginosa infections in patients, hospital means, and personnel’s
specimens. J Res Med Sci 17(4), 332-7.
www.cdc.gov/media/releases/2013/p0305_deadly_bacteria.html
www.medicalnewstoday.com/articles/197616.php
Huang CR, Lu CH, Chuang
YC, Tsai NW, Chang CC, Chen SF, Wang HC, Chien CC, Chang WN. 2007
“Adult Pseudomonas aeruginosa meningitis: high incidence of
underlying medical and/or post neurosurgical conditions and high mortality
rate.” Jpn J Infect Dis 60(6), 397-9.
Kashyap A, Gupta R,
Sharma R, Verma VV, Gupta S, et al. 2017. New Delhi Metallo Beta
Lactamase: Menace and its Challenges. J Mol Genet Med 11, 299
doi:10.4172/1747-0862.1000299
Khan AU, Maryam L,
Zarrilli R. 2017. Structure, Genetics and Worldwide Spread of New Delhi
Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol 17, 101.
Lee K, Lim YS, Yong D,
Yum JH, Chong Y. 2003. “Evaluation of the Hodge test and the Imipenem-EDTA
double-disk synergy test for differentiating of metallo-β- lactamase-producing
clinical isolates of Pseudomonas spp. and Acinetobacter Spp.” J Clin
Microbiol 41(10), 4623-9.
Livermore DM, Woodford
N. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol 3, 489-495.
Murray Patrick R, Baron
Ellen Jo, Jorgensen James H, Landry Marie Louise. 2003. Manual of clinical
Microbiology, Vol I, 8th edition, ASM Press 734-834.
Nordmann P, Poirel L,
Carrer A, Toleman MA, Walsh TR. 2011. How to detect NDM-1
producers. J Clin Microbiol 49, 718-21.
Obritsch MD, Fish DN,
MacLaren R, Jung R. 2005. Nosocomial infections due to multidrug-resistant
Pseudomonas aeruginosa: epidemiology and treatment options.
Pharmacotherapy 25(10), 1353-64.
Padhi S. 2011. New
Delhi metallo-beta-lactamase: A weapon for the newly emerging drug-resistant
bacteria. Indian J Med Sci 65, 317–320.
Pitout JD1, Gregson DB,
Poirel L, McClure JA, Le P, Church DL. 2005. Detection of Pseudomonas
aeruginosa producing metallo-beta-lactamases in a large centralized laboratory.
J Clin Microbiol 43(7), 3129-35.
Raghunath D. 2010.
New metallo β-lactamase NDM-1. Indian J Med Res 132(5), 478-81.
Rubin SJ, Granato PA,
Wasilauskas BL. 1985. Glucose-nonfermenting gram-negative bacteria p.
330-349. In Lennette EH, Balows A, Hausler WJJr, Shadomny HJ (ed.), Manual of
clinical microbiology, 4th ed. American Society for Microbiology, Washington,
D.C.
Shanthi M, Sekar U,
Kamalanathan A, Sekar B. 2014. Detection of New Delhi metallo beta
lactamase-1 (NDM-1) carbapenemase in Pseudomonas aeruginosa in a
single centre in southern India. Indian J Med Res. 140(4), 546-50.
Washington Winn Jr,
Allen Stephen, Janda William, Koneman Elmer, Procop Gary, Screckenberger Paul,
Woods Gail. 2006. “The non-fermentative Gram negative bacilli” Koneman’s
Colour atlas and textbook of diagnostic Microbiology, 6th edition,
Lippincott William & Wilkins 303-392.
Watanabe M, Iyobe S,
Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas
aeruginosa. Antimicrob. Agents Chemother 35, 147–151.
Winn W Jr, Allen S,
Janda W, Koneman E, Procop G, Schreckenberger P, et al. 2006.
editors. In: Koneman’s Color Atlas and textbook of Diagnostic Microbiology. 6th
ed. USA: Lippincott Williams and Wilkins Company;. Nonfermenting Gram negative
bacilli; pp. 305-91.
Yong D, Lee K, Yum JH,
Shin HB, Rossolini GM, Chong Y. 2002. “Imipenem-EDTA disk method for
differentiation of metallo-beta-lactamase-producing clinical isolates of
Pseudomonas spp. and Acinetobacter spp.” J Clin Microbiol 40(10), 3798-801
Yong D, Toleman MA,
Giske CG, Cho HS, Sundman K, Lee K, et al. 2009. Characterization of
a New Metallo-β-lactamase gene bla and a novel erythromycin esterase gene
carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14
from India. Antimicrob Agents Chemother 53, 5046–54.











%20in%20full.JPG)


0 comments:
Post a Comment